School Resources
The Jalview team has taken advantage of Jalview’s easy-to-use interface to develop a variety of materials suitable for schools and home learning. The exercises are designed to let pupils view and interact with a selection of biological molecules. Encouraging pupils to explore fundamental biological concepts in an independent manner.
Jalview in Schools Workbook
The ‘Visualising DNA, RNA & Proteins’ workbook contains 4 easy-to-run web-based bioinformatics projects that are aimed at 15-18 year-old secondary school pupils studying biology at Higher, Advanced Higher or A levels.
Project 1: single click exercises to view DNA & RNA sequences and 3D structures.
Project 2: single click exercise to view human myoglobin protein sequence and 3D structure, followed by an exercise that uses a similarity tree to compare the myoglobin sequences from different animals.
Project 3: single click exercises to view the sequences and 3D structures of proteins with a range of different biological functions.
Project 4: exercise to view the exons and introns in the HBB gene, and an exercise to identify the genetic mutation on the DNA that is linked to sickle cell anaemia, and its location on the 3D structure of haemoglobin protein.
IT Requirements: A computer with a web browser and internet access. Since JalviewJS opens in a web browser window, no software has to be downloaded.

View the Workbook
Click the link to view the workbook in an adjacent window (or download depending on the browser), then follow the instructions to run the exercises.
Links to Run the Exercises
Clicking the links below will start each exercise by opening JalviewJS in an adjacent browser tab and reading in files as appropriate. Note that it may take a few seconds to load the sequences.
Project 1
Exercise 1: Click to view a fragment of DNA
Exercise 2: Click to view the coding sequence of DNA and codons alongside the protein product
Exercise 3: Click to view an RNA molecule
Project 2
Exercise 4: Click to view Human Myoglobin protein sequence and 3D structure
Exercise 5: Click to view Myoglobin protein sequences from different species and create a Tree
Project 3
Exercise 6: View protein sequences and 3D structures
-
Click to view Haemoglobin protein (transport)
-
Click to view Collagen protein (structure)
-
Click to view Amylase protein (enzyme)
-
Click to view Antibody protein (defence)
-
Click to view Insulin protein (signalling)
-
Click to view Ferritin protein (storage)
-
Click to view Myoglobin protein (transport)
Project 4
Exercise 7: Click to view Exons & Introns in the DNA of the HBB gene
Exercise 8: Click to view the coding DNA of the HBB gene & its protein product
Additional Exercises
Advanced Exercise 1: Click to open the JalviewJS desktop window in an adjacent browser window
Advanced Exercise 2: Click to view the coding DNA of the HBB gene & its protein product - reading files from public biological databases (EMBLCDS ID = CAG46711)
Frequently Asked Question - I’m having trouble viewing the 3D structure
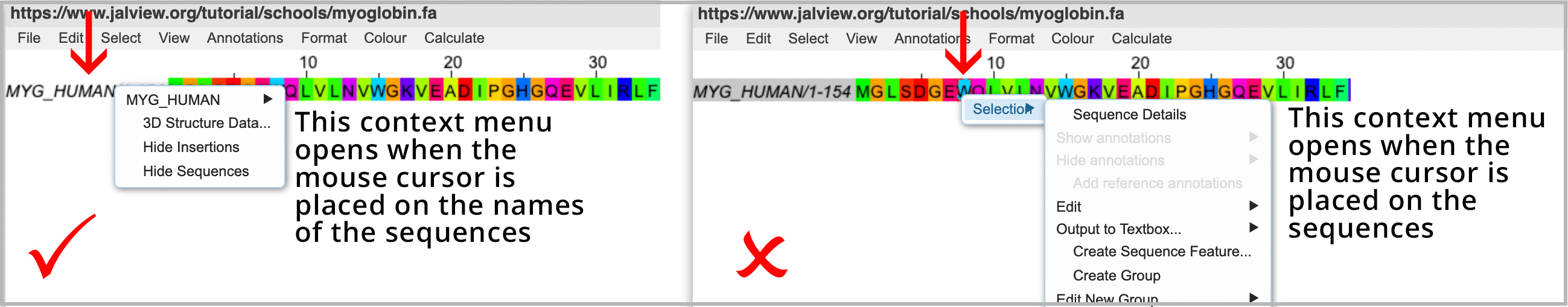
Loading the 3D structure involves opening a context menu. The position of the mouse is critical when opening context menus. Click this Jalview School Video 2 link to view the manoeuvre.
- Place the mouse over the name of the sequences (not the sequences themselves)
- Then right-click the mouse to open the context menu
Jalview School Videos
(i) Why do we need bioinformatics software such as Jalview?
(ii) Introducing the Jalview Schools Workbook
(iii) What is Jalview?
Jalview is one of the most widely used applications for viewing and analysing DNA, RNA and protein sequences. It is free-to-use computer software with an interactive multi-window interface. This allows scientists to read files directly from public biological databases and view sequences, 3D structures and evolutionary trees. Jalview has a number of analysis tools for aligning sequences, measuring similarities, exploring features and comparing structures. Jalview has been developed in Prof Geoff Barton’s group in the School of Life Sciences at the University of Dundee, with funding from the BBSRC and the Wellcome Trust.
Read the RCSB PDB Newsletter article featuring the Jalview School Workbook

Free Public Biological Databases
(i) UniProt is a database of protein sequences (https://www.uniprot.org).
(ii) Protein Data Bank (PDB) is a database of crystallographic, three-dimensional structural data of large biological molecules (https://www.rcsb.org).
(iii) Ensembl is a genomic database (https://ensemblgenomes.org).
(iv) EMBL (CDS) data originates from the European Nucleotide Archive (ENA) database of annotated DNA and RNA sequences (https://www.ebi.ac.uk/ena).
(v) PFAM is a database of protein families (https://pfam.xfam.org/).
(vi) RFAM is a database containing information about non-coding RNA families and other structured RNA elements (https://rfam.xfam.org).
Database Identifiers
PDB IDs used in the workbook
| PDB ID | Molecule | Function | Link |
|---|---|---|---|
| 3DSE | DNA fragment | https://pdb101.rcsb.org/motm/23 | |
| 2GIS | RNA | https://pdb101.rcsb.org/motm/15 | |
| 1smd | Amylase | Enzyme | https://pdb101.rcsb.org/motm/74 |
| 1cag | Collagen | Structure | https://pdb101.rcsb.org/motm/7 |
| 1igt | Antibody | Defence | https://pdb101.rcsb.org/motm/21 |
| 5xb1 | Ferritin | Storage | https://pdb101.rcsb.org/motm/35 |
| 6bcx | Insulin | Signalling | https://pdb101.rcsb.org/motm/14 |
| 1mbn | Myoglobin | Transport | https://pdb101.rcsb.org/motm/1 |
Uniprot Myoglobin IDs used in the workbook
| Uniprot ID | Species |
|---|---|
| P02144 | Human |
| P02147 | Gorilla |
| P02185 | Whale |
| P02192 | Cow |
| P04247 | Mouse |
| P68276 | Dolphin |
| P02197 | Chicken |
| P63113 | Dog |
| P02190 | Sheep |
| P02187 | Elephant |
P02144; P02147; P02185; P02192; P04247; P68276; P02197; P85077; P63113; P02190; P02187
DNA IDs used in the workbook
CAG46747 EmblCDS ID for MB gene that codes for myoglobin protein P02144.
CAG46711 EmblCDS ID for HBBgene that codes for Hemoglobin subunit beta protein P68871.
Acknowledgements
The workbook was produced by Suzanne Duce with help from Mungo Carstairs, Benedict Soares, Bob Hanson, Dmitry Finkelbergs, Charlotte Campbell, Jim Procter and Geoff Barton.